Will Bolton
AI for Health PhD Student
Imperial College London
Will is a PhD researcher in AI for healthcare at Imperial College London. His work focuses on machine learning based clinical decision support for antimicrobial optimisation. To date he has published 5 first authored publications in leading medical journals and AI conferences and conducted an internship at GSK.ai on the reliability of LLM’s in the biomedical domain. Beyond research he is highly interested in startups and how new innovative technology can create solutions to real world problems. He is currently part of EWOR as a Pre-Idea Fellow and ConceptionX as a venture scientist to explore entrepreneurship opportunities post his PhD.
- Artificial Intelligence
- Healthcare
- Entrepreneurship
PhD in Artificial Intelligence, 2024
Imperial College London
MPhil Bioscience Enterprise, 2019
University of Cambridge
BSc in Biochemistry, 2018
Imperial College London
Experience
Featured Publications
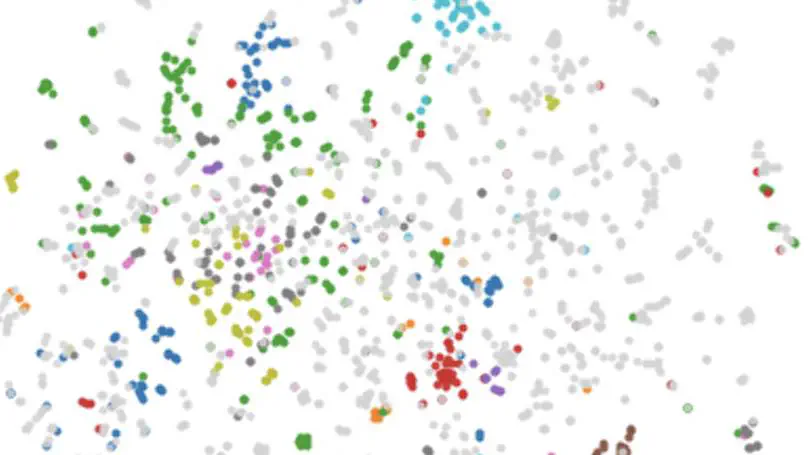
Co-morbidities or long-term medical conditions are an essential piece of clinical information that can drastically alter a patient’s treatment, management and outcome. However, such data is difficult to apply to artificial intelligence (AI) systems due to its heterogeneity, sparsity and combinatorial complexity. This text proposes a novel pipeline to address such pitfalls by utilizing the structure of Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), the most extensive clinical vocabulary in the world, to create disease and co-morbid patient embeddings which can be used in downstream AI applications. We demonstrate that our method- ologies outperform existing approaches in classification and similar patient retrieval tasks. This research highlights that appropriate extraction and representation of clinical knowledge through innovative approaches can enable AI systems to advance personalized healthcare.
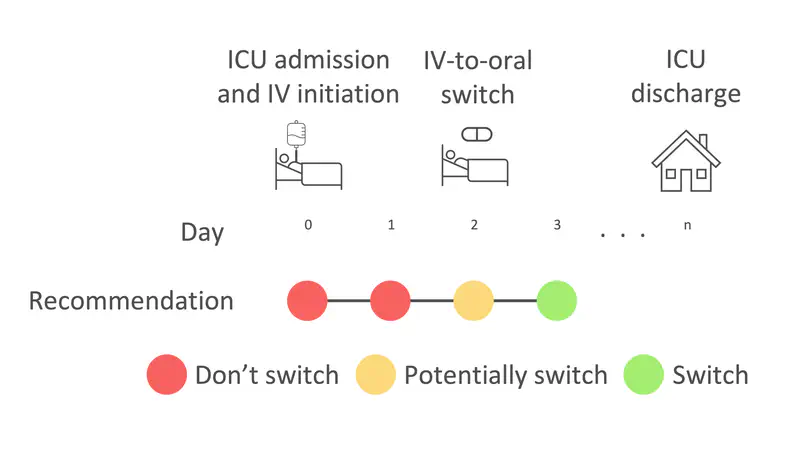
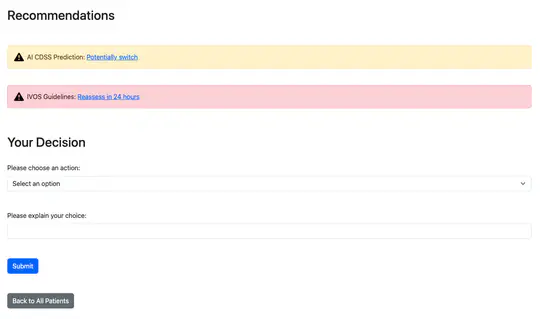
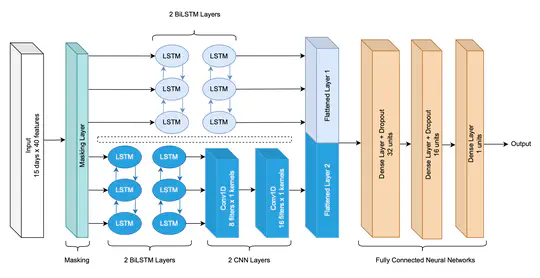
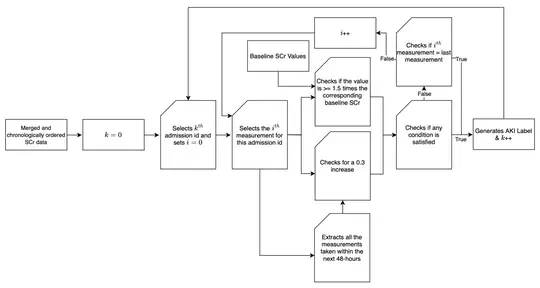
Antimicrobial resistance (AMR) and healthcare associated infections pose a significant threat globally. One key prevention strategy is to follow antimicrobial stewardship practices, in particular, to maximise targeted oral therapy and reduce the use of indwelling vascular devices for intravenous (IV)administration. Appreciating when an individual patient can switch from IV to oral antibiotic treatment is often non-trivial and not standardised. To tackle this problem we created a machine learning model to predict when a patient could switch based on routinely collected clinical parameters. 10,362 unique intensive care unit stays were extracted and two informative feature sets identified. Our best model achieved a mean AUROC of 0.80 (SD 0.01) on the hold-out set while not being biased to individuals protected characteristics. Interpretability methodologies were employed to create clinically useful visual explanations. In summary, our model provides individualised, fair, and interpretable predictions for when a patient could switch from IV-to-oral antibiotic treatment. Prospectively evaluation of safety and efficacy is needed before such technology can be applied clinically.
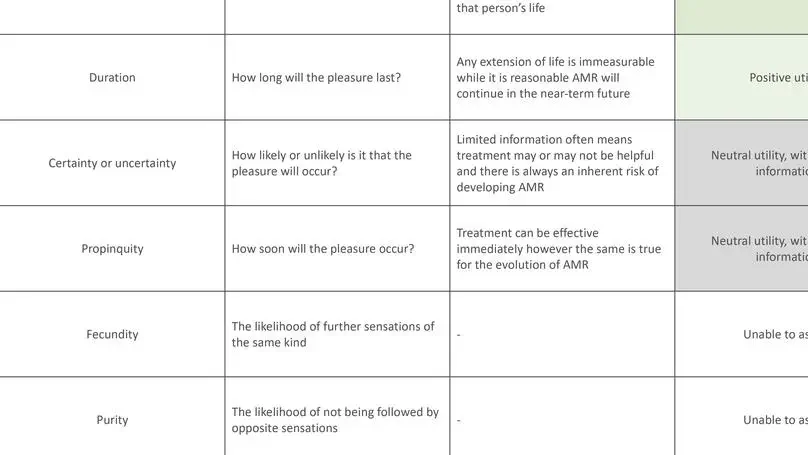
The use of decision-support systems based on artificial intelligence approaches in antimicrobial prescribing raises important moral questions. Adopting ethical frameworks alongside such systems can aid the consideration of infection-specific complexities and support moral decision-making to tackle antimicrobial resistance.
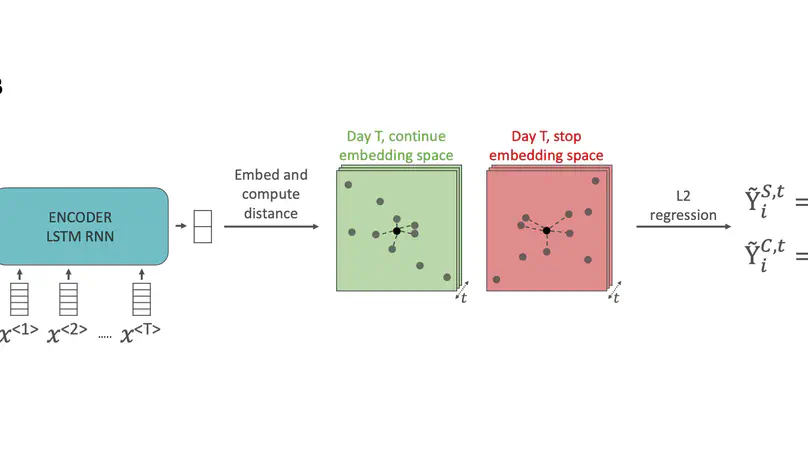
The decision on when it is appropriate to stop antimicrobial treatment in an individual patient is complex and under-researched. Ceasing too early can drive treatment failure, while excessive treatment risks adverse events. Under- and over-treatment can promote the development of antimicrobial resistance (AMR). We extracted routinely collected electronic health record data from the MIMIC-IV database for 18,988 patients (22,845 unique stays) who received intravenous antibiotic treatment during an intensive care unit (ICU) admission. A model was developed that utilises a recurrent neural network autoencoder and a synthetic control-based approach to estimate patients’ ICU length of stay (LOS) and mortality outcomes for any given day, under the alternative scenarios of if they were to stop vs. continue antibiotic treatment. Control days where our model should reproduce labels demonstrated minimal difference for both stopping and continuing scenarios indicating estimations are reliable (LOS results of 0.24 and 0.42 days mean delta, 1.93 and 3.76 root mean squared error, respectively). Meanwhile, impact days where we assess the potential effect of the unobserved scenario showed that stopping antibiotic therapy earlier had a statistically significant shorter LOS (mean reduction 2.71 days, p-value <0.01). No impact on mortality was observed. In summary, we have developed a model to reliably estimate patient outcomes under the contrasting scenarios of stopping or continuing antibiotic treatment. Retrospective results are in line with previous clinical studies that demonstrate shorter antibiotic treatment durations are often non-inferior. With additional development into a clinical decision support system, this could be used to support individualised antimicrobial cessation decision-making, reduce the excessive use of antibiotics, and address the problem of AMR.